Hacking
Florian Ziemen
Tobias Kölling
Lukas Kluft
2025-05-12
Have fun
and
learn about Earth
Smooth workflows for km-scale data
(Code)
Made possible by
Loading less data
Load only the data you need
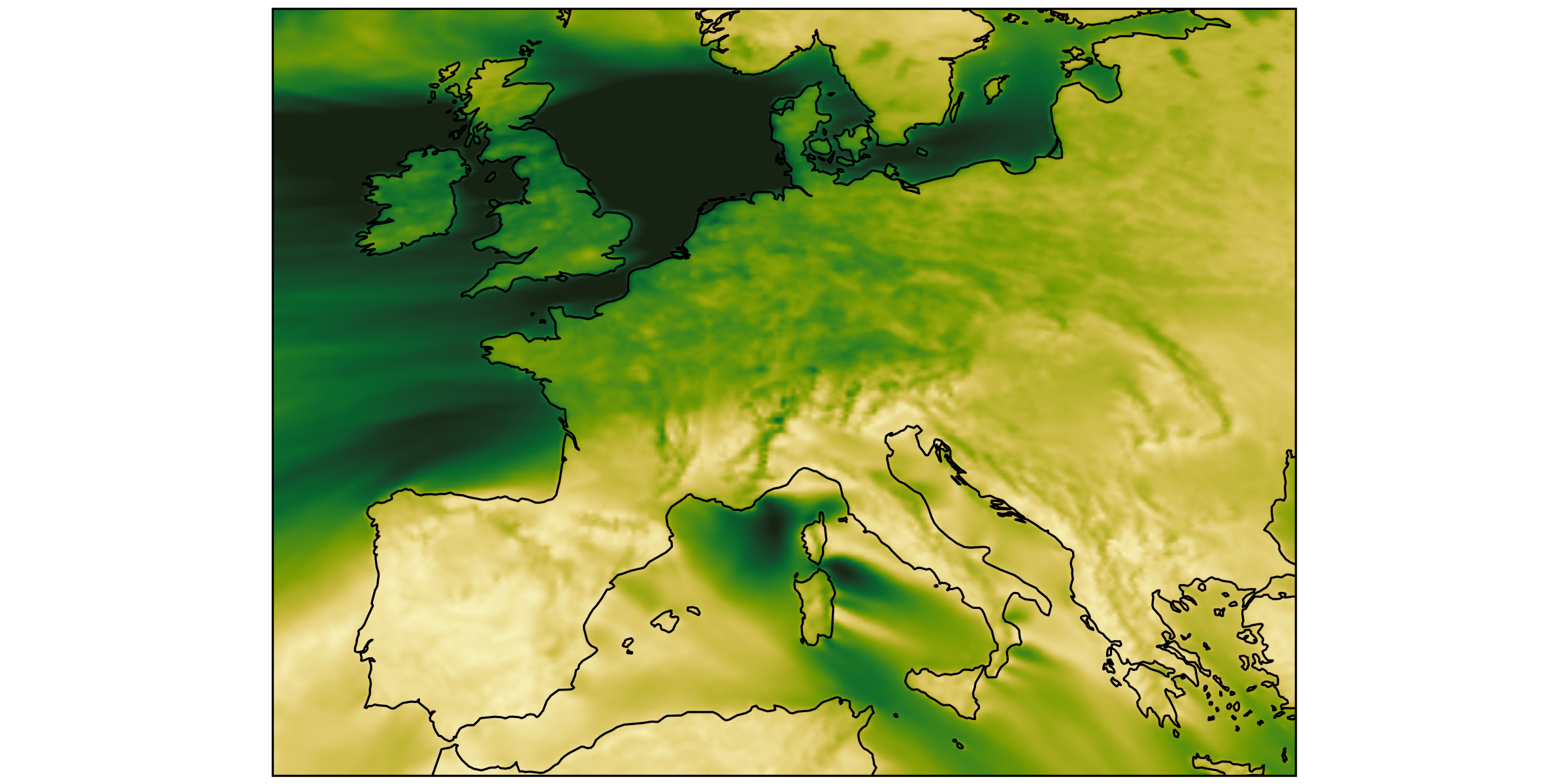
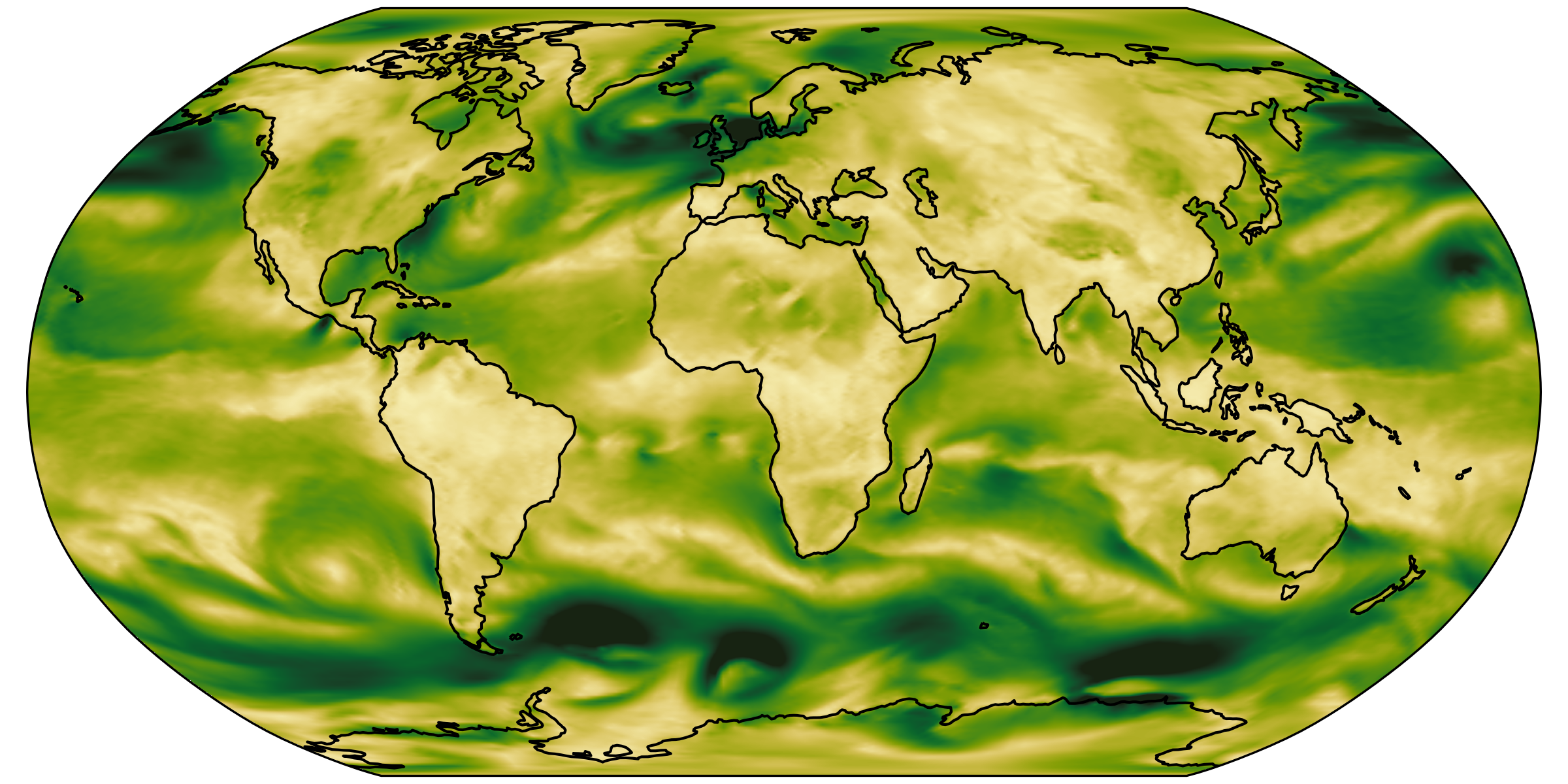
Both plots have the same number of pixels.
You should load the same amount of data to make them.
Our approach
- HEALPix Grid
- Resolution hierarchies
- Chunked storage (you’ll probably not see much of this)
- Catalogs grouping the datasets
HEALPix
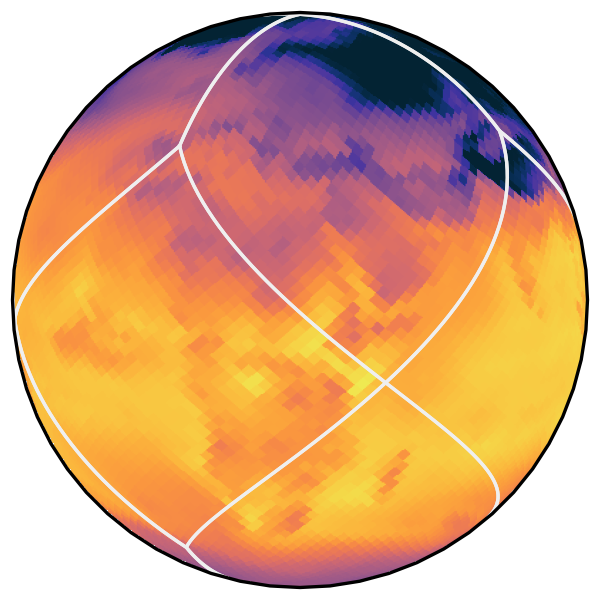
HEALPix features
- Uniform coverage of Earth
- Direct translation between lat/lon and pixel ID
- Cells arranged in isolatitude bands
- Index is a space-filling curve

Healpix hierarchy
Refinement by splitting each cell into four finer cells
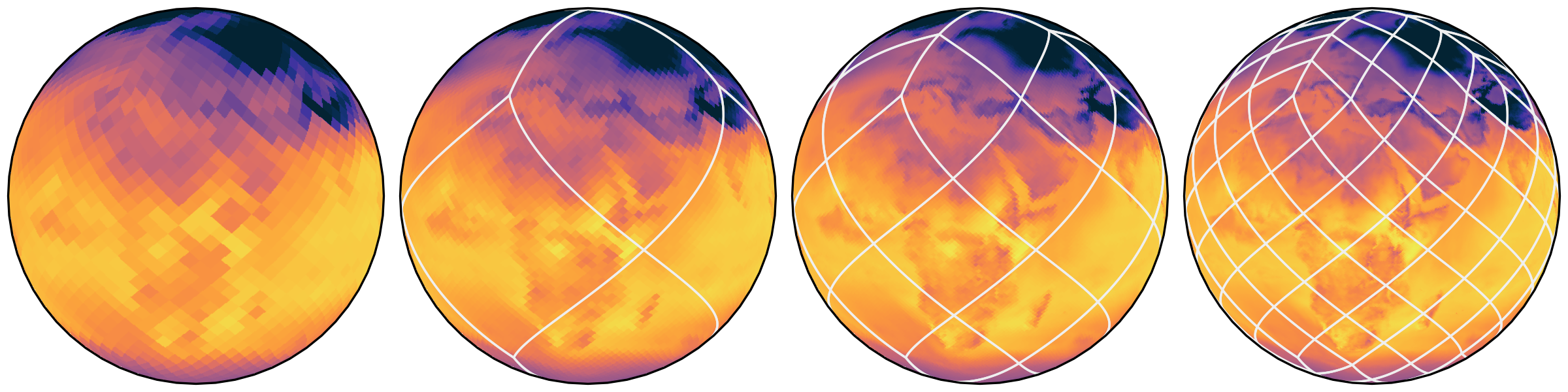
Load only the data you need
- Global mean -> Level 0 (12 cells)
- Test a global map -> Level 5 (12288 cells)
- Fill a screen -> Level 9 (3M cells)
- Analyze a detail -> load only the region
Catalogs and phone books
- Basically your phone’s list of contacts
- You ask for a name, it will call the dataset
- Update it when the phone number of your contact changes
- No update needed when your contact gets a new phone
- share it in the team, and save effort updating
Catalogs
Call it by its name, not by the location
cat["casesm2_10km_cumulus"].to_dask()instead of
xr.open_dataset("/lustre/persons_name/experiments/attempt7/outdata/data_*_74_b.nc")No need to know where data is.
Parameterize variants
cat["casesm2_10km_cumulus"](zoom=5, time="PT3H")
Let’s get real
Working with the catalog
Index generated from the catalog
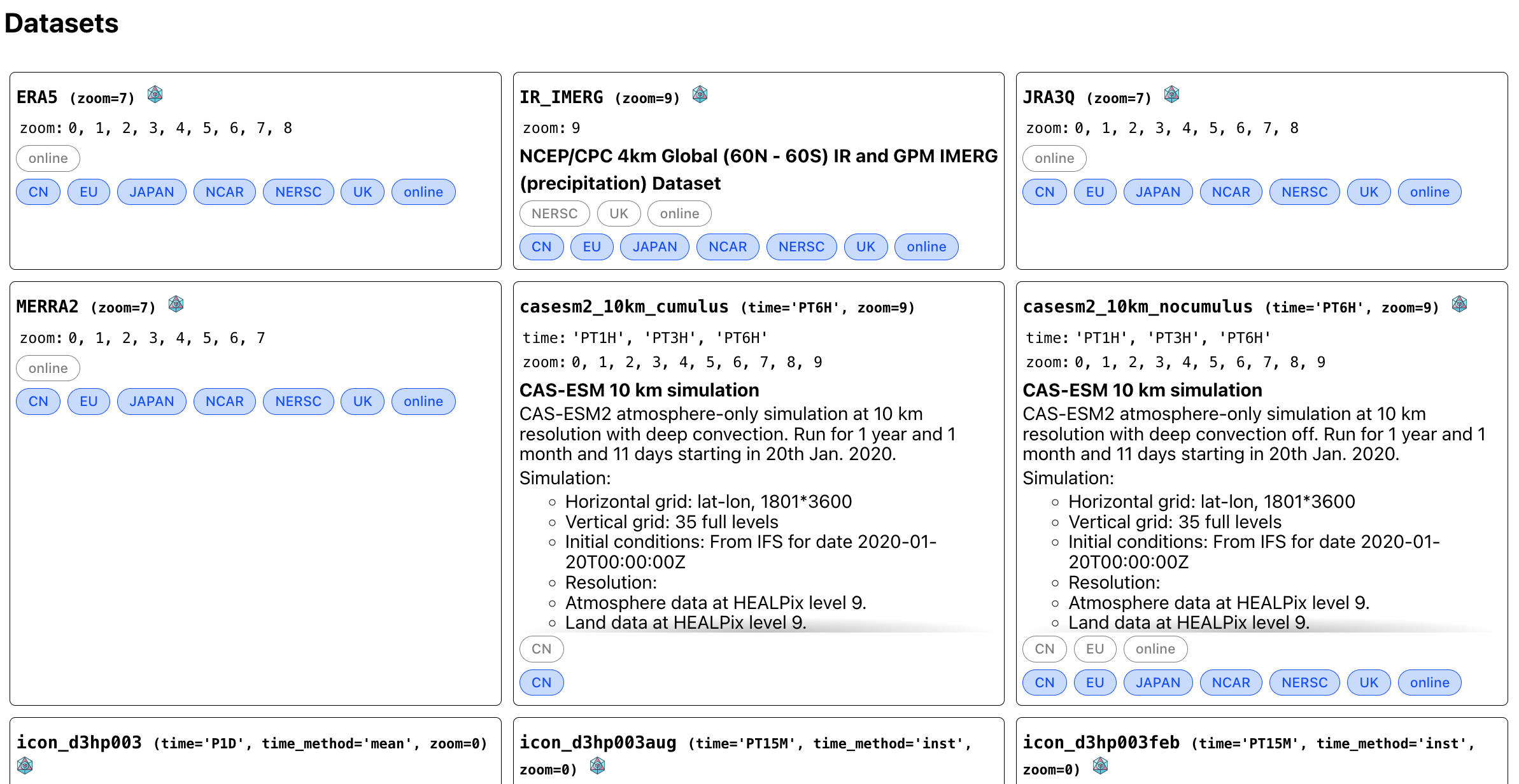
Locations and datasets
- Our catalog has two dimensions
- Location
- Dataset
- Any dataset can be hosted at different locations
- Online datasets are availabe at all locations
- Local copies are preferred
Load the catalog
Get the phone number of a dataset
'/data2/share/florain/CAS-ESM2_10km_cumulus_3d6h_z9.zarr'Call the dataset directly
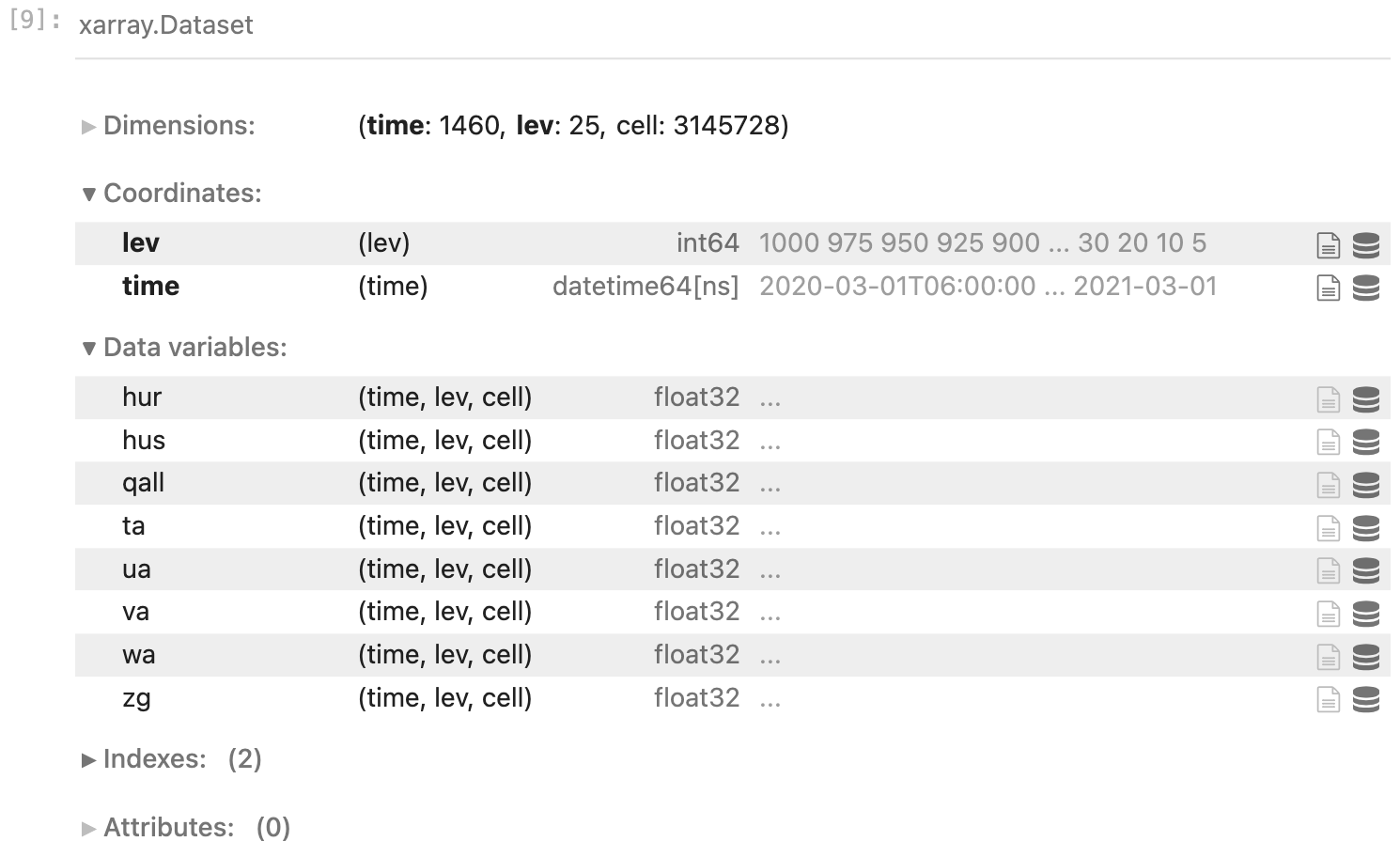
Look at the possible parameters
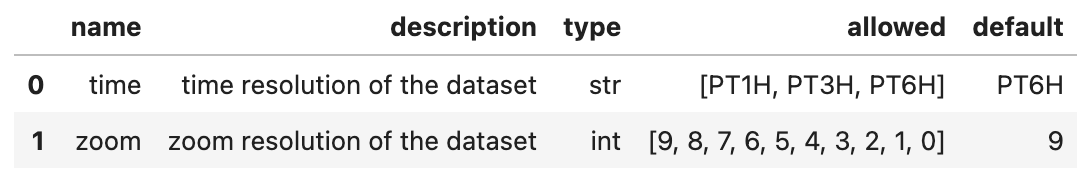
Load a specific variant
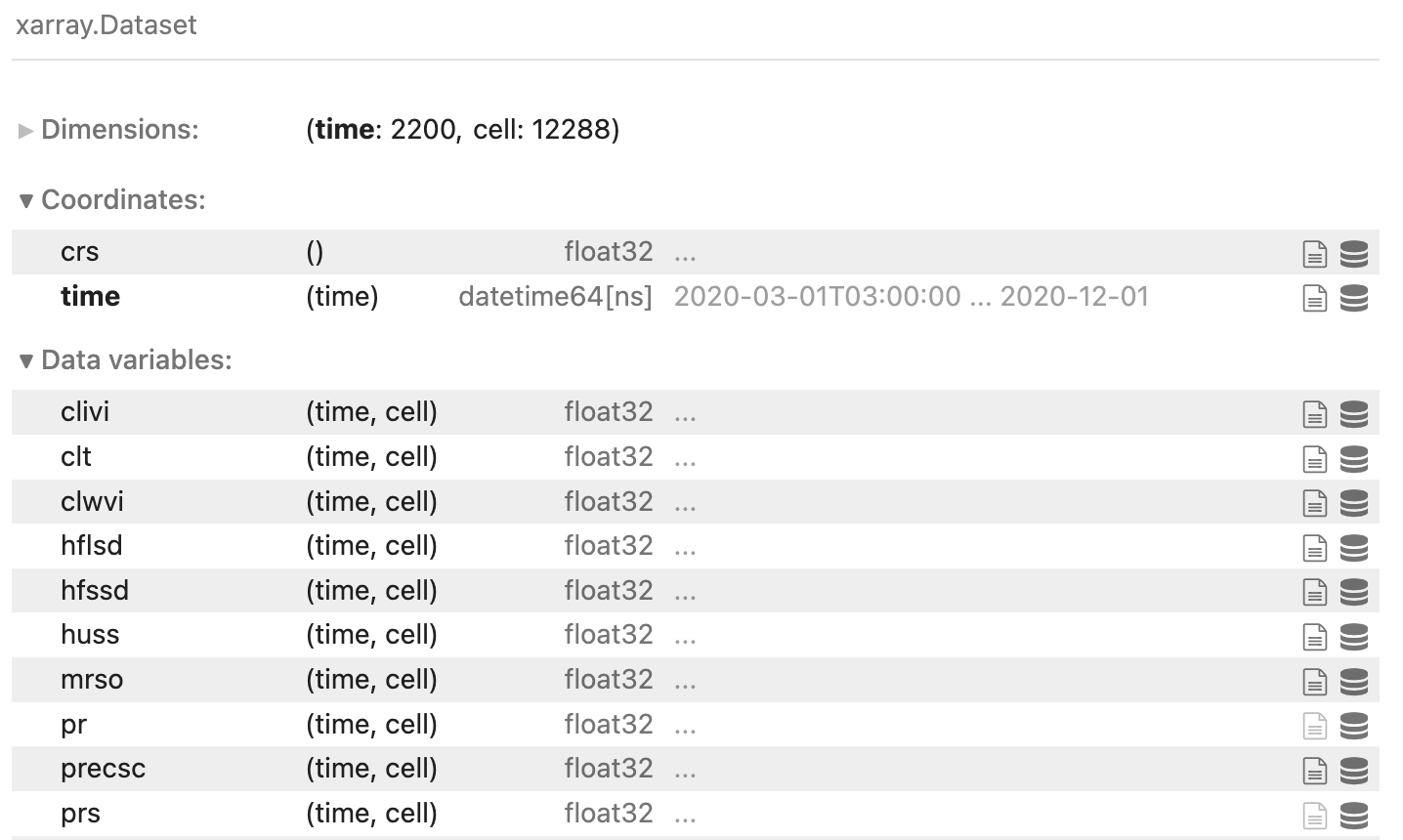 # Mapmaking
# Mapmaking
A simple world map
import easygems.healpix as egh
var = "tas"
plot_time = "2020-05-12T09:00:00"
cmap = "inferno"
egh.healpix_show(ds[var].sel(time=plot_time), cmap=cmap) 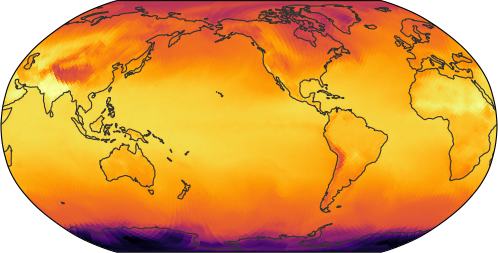
increasing the resolution
ds = cat[name](zoom=7, time="PT3H").to_dask()
egh.healpix_show(ds[var].sel(time=plot_time), cmap=cmap) 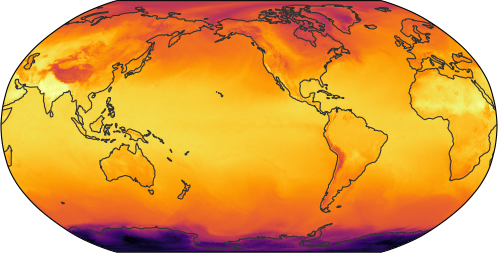
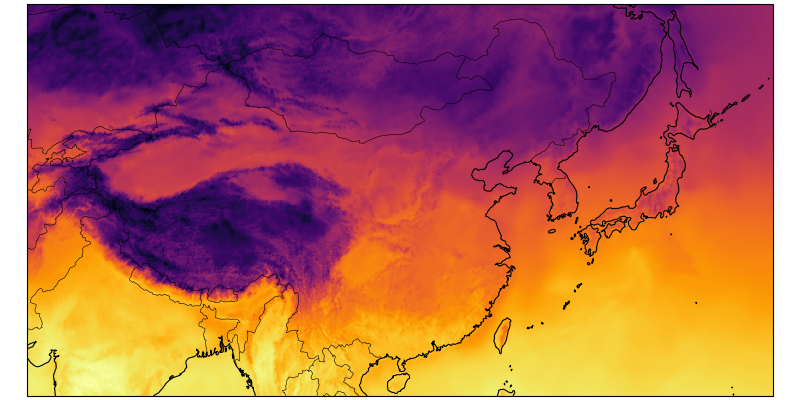
Zooming in
import cartopy.crs as ccrs
import cartopy.feature as cf
ds = cat[name](zoom=9, time="PT3H").to_dask()
projection = ccrs.Robinson(central_longitude=120)
fig, ax = plt.subplots(
figsize=(8, 4),
subplot_kw={"projection": projection},
constrained_layout=True
)
ax.set_extent([70, 150, 18, 55], crs=ccrs.PlateCarree())
egh.healpix_show(ds.tas.isel(time=0),
ax=ax,
cmap=cmap)
ax.add_feature(cf.COASTLINE, linewidth=0.8)
ax.add_feature(cf.BORDERS, linewidth=0.4)Zooming in
More map examples on easy.gems
https://easy.gems.dkrz.de/Processing/healpix/healpix_cartopy.html
Zonal means
ds = cat[name](zoom=5, time="PT3H").to_dask().pipe(egh.attach_coords)
pr = ds['pr'].mean(dim='time').groupby(ds.lat).mean()
pr.plot()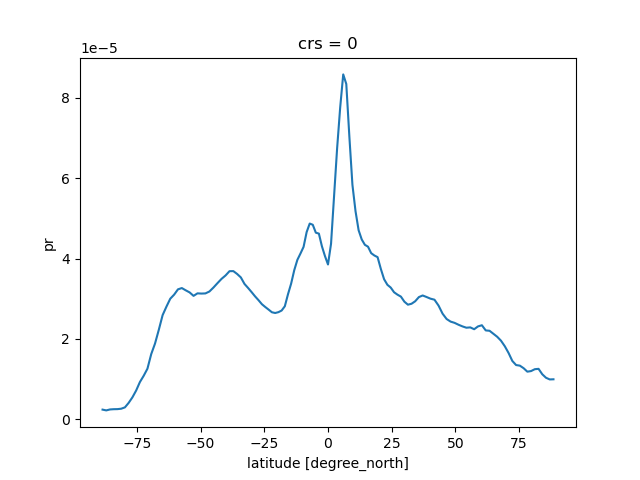
Zonal section
import matplotlib.pyplot as plt
ds = cat[name](zoom=5, time="PT6H").to_dask().pipe(egh.attach_coords)
ua = ds['ua'].mean(dim='time').groupby(ds.lat).mean()
ua.plot()
plt.ylim(plt.ylim()[::-1])
plt.title (f"Zonal mean zonal wind speed (m/s)")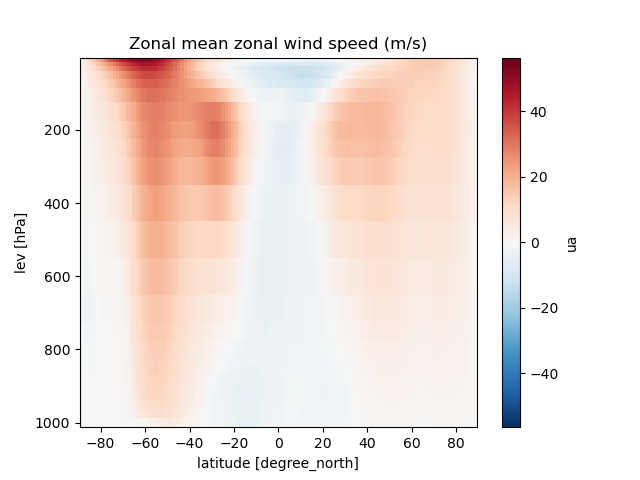
Space-time diagram
ds = cat[name](zoom=7, time="PT3H").to_dask().pipe(egh.attach_coords)
Slim, Nlim = -15.0, 35.0
pr = (
ds['pr']
.where((ds["lat"] > Slim) & (ds["lat"] < Nlim), drop=True)
.groupby("lat")
.mean()
).coarsen(time=8).mean().transpose().compute()
pr.plot(cmap="Blues", vmax=0.0001)
plt.title(f"zonal mean precipitation (kg m-2 s-1)")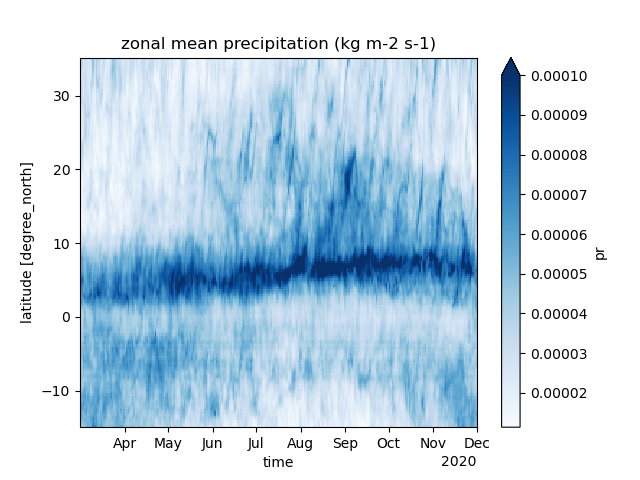
More on time-space diagrams
https://easy.gems.dkrz.de/Processing/healpix/time-space.html
黑客马拉松圆满成功
A few words on datasets and Catalogs
Build clean datasets
- All variables together that fit together
- Cutting parts out is easier than gluing together
- Time spent on making them clean is saved during analysis
- Variants for coarsening should follow a logic
Catalogs arrange datasets logically
- Group dataset by topic
- Tree-Style catalogs allow for nesting
- Try to have exactly one catalog with good location
- Catalogs can be nested
